Triticeae Toolbox / WheatCAP Prediction Competition
We have set up a mailing list for those participating in the Prediction Challenge. We will use this list to send occasional updates about new data added to the database or tools that may be relevant for the challenge.
To sign up, go to the T3 Mailing List Subscription Form, enter your name and email address, and make sure the T3 Prediction Challenge list is selected.
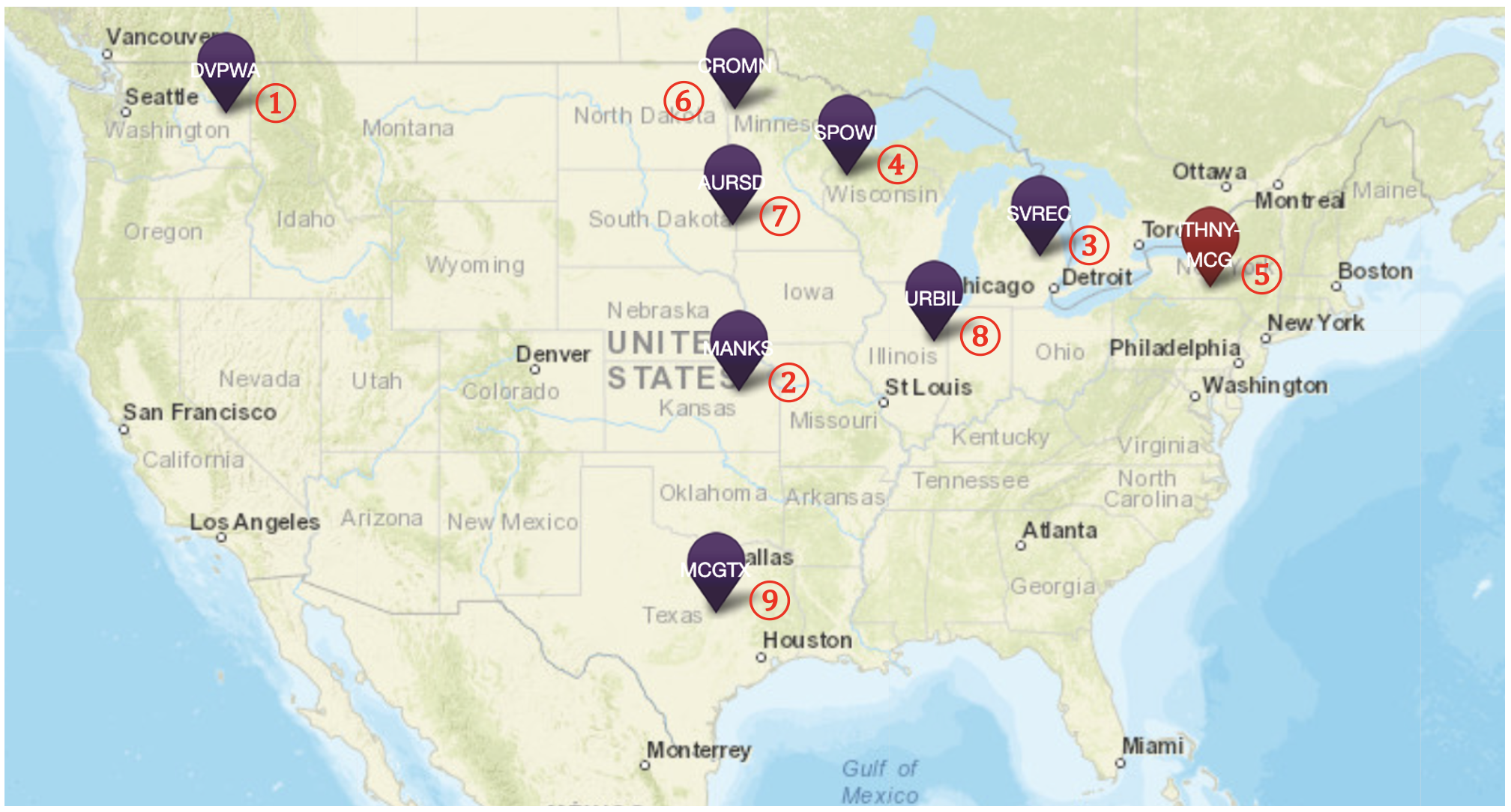
The objective will be to use data available on The Triticeae Toolbox (T3/Wheat) to predict yield (Grain yield - kg/ha|CO_321:0001218) performance of wheat accessions in 9 separate test trials across the USA. Important metadata for these trials is on T3/Wheat, but the phenotypes measured in them are not. For all of these trials, genotypic data is available, though it is somewhat messy: 1. Not all accessions in the trial are genotyped; 2. Different genotyping protocols have been used in different trials.
The names of the 9 trials are:
- AWY1_DVPWA_2024
- TCAP_2025_MANKS
- 25_Big6_SVREC_SVREC
- OHRWW_2025_SPO
- CornellMaster_2025_McGowan
- 24Crk_AY2-3
- 2025_AYT_Aurora
- YT_Urb_25
- STP1_2025_MCG
Contestants will submit a folder containing predictions for genotyped entries in the trials for "CV0" and "CV00" cases (Jarquin et al. 2017; see below). The winning algorithm will be judged on the basis of the highest average prediction accuracy across trials and cross validation cases.
Training Data
We have identified T3 data that could be reasonably applied to predicting the Predictathon trials using simple heuristics:
- We identified phenotyping trials that evaluated some of the same accessions as each Predictathon trial. The assumption is that the overall germplasm in those trials will be related to the Predictathon accessions.
- We identified archived VCFs with genotype data from a minimum number of each of the accessions in the putative training trials.
Below are links to the data and metadata that could be used as training sets for each of the prediction trials:
| Study Name | Study ID | Training Data |
|---|---|---|
| AWY1_DVPWA_2024 | 10678 | Download |
| TCAP_2025_MANKS | 10680 | Download |
| 25_Big6_SVREC_SVREC | 10675 | Download |
| OHRWW_2025_SPO | 10679 | Download |
| CornellMaster_2025_McGowan | 10676 | Download |
| 24Crk_AY2-3 | 10674 | Download |
| 2025_AYT_Aurora | 10673 | Download |
| YT_Urb_25 | 10677 | Download |
| STP1_2025_MCG | 10681 | Download |
This data was generated from scripts found in this GitHub repository:
https://github.com/jeanlucj/T3_predictathon_find_training_trials
Genotype Data for Prediction Trials
Below is a table listing the best genotype projects and protocols (in this case, having the highest number of genotyped accessions from the trial) on T3 for each prediction trial.
| study_name | project_name | project_id | protocol_name | protocol_id | download |
|---|---|---|---|---|---|
| 2025_AYT_Aurora | SDSU_2025_GBS | 11050 | GBS SDSU 2025 | 301 | Download Archived VCF File |
| 24Crk_AY2-3 | UMN_2022_GBS | 10671 | GBS UMN 2022 | 294 | Download Archived VCF File |
| 25_Big6_SVREC_SVREC | BIGSIX_2024_AgriSeq_4K | 10684 | ThermoFisher AgriSeq 4K | 296 | Download Archived VCF File |
| CornellMaster_2025_McGowan | Cornell_WWMasterScreening_2024 | 11049 | GBS Cornell 2024 | 300 | Download Archived VCF File |
| YT_Urb_25 | UIUC_2024_GBS_V2 | 11054 | GBS UIUC 2024 | 302 | Download Archived VCF File |
| AWY1_DVPWA_2024 | WSU_2023_GBS | 14512 | GBS WSU 2023 | 311 | Download Archived VCF File |
| OHRWW_2025_SPO | UWM_2023_3K | 11009 | Wheat 3K | 299 | Download Archived VCF File |
| TCAP_2025_MANKS | KSU_2024_Allegro_V2 | 10830 | Allegro V2 | 297 | Download Archived VCF File |
| STP1_2025_MCG | TAMU_2025_MCG25 | 11052 | GBS TAMU MCG25 | 303 | Download Archived VCF File |
NOTE: New data (in particular genotypic data) will be posted to T3/Wheat during the contest period. It will be valuable to recheck before the competition closes to assemble final training data.
A presentation describing useful methods of programmatic access to T3 to facilitate algorithm development will be given at PAG in the Breedbase Workshop Sunday, January 11, 2026: https://pag.confex.com/pag/33/meetingapp.cgi/Session/14070
The competition will close on Friday, March 13th, 2026
The deadline to submit predictions has been extended to Friday, March 27th.
An upload link to submit a zipped folder containing the predictions will be posted here prior to that date.
Details
Descriptions of CV0 and CV00 cross validation scenarios (Jarquin et al. 2017).
Jarquín, D., C. Lemes da Silva, R.C. Gaynor, J. Poland, A. Fritz, et al. 2017. Increasing genomic-enabled prediction accuracy by modeling genotype × environment interactions in Kansas wheat. Plant Genome 10(2): plantgenome2016.12.0130. doi: 10.3835/plantgenome2016.12.0130.
For CV0, for the purpose of making predictions, the function should exclude the focal trial itself from the training data but may use all data in any other trial present in the database, including data on the entries in the focal trial. For CV00, the function should exclude the focal trial and any observations on entries in the focal trial occurring in other trials from the training data prior to making predictions. For each prediction task, accuracy will be calculated as the correlation between prediction and observed phenotypes over all genotyped accessions in the trial. Average accuracy will be calculated across 18 prediction tasks (9 separate test trials; CV0 and CV00 accuracy for each trial).
Submission Requirements
Folder Structure:
The zipped folder submitted should contain one methods description text file and 9 sub-directories, each named with the trial name for which the sub-directory contains predictions. For example, if the sub-directory contains predictions for the trial "AYT_Timbuktu24", then the sub-directory should be named "AYT_Timbuktu24". Each sub-directory should contain six csv files. Continuing the example, the sub-directory should contain:
- CV0_Predictions.csv
- CV0_Trials.csv
- CV0_Accessions.csv
- CV00_Predictions.csv
- CV00_Trials.csv
- CV00_Accessions.csv
These files should be formatted as follows.
CSV file formats:
- CV0_Predictions.csv and CV00_Predictions.csv: csv file with two columns, "germplasmName" and "prediction". All accessions in the focal trial that are genotyped should have a prediction.
- CV0_Trials.csv and CV00_Trials.csv: a csv file with a single column, "studyName", containing the trial names of the trials used for training the prediction model.
- CV0_Accessions.csv and CV00_Accessions.csv: a csv file with a single column, "germplasmName", containing the accession names used for training the prediction model.
Methods description: Common sections (these sections can be brief)
This should be a text, rtf, gdoc, docx, or pdf file.
- URL to a publicly accessible code repository (e.g., github.com or bitbucket.org) that contains the prediction algorithm.
-
Data retrieval method: programmatic access or GUI download
- If the former, a list of the BrAPI calls used
- If the latter, the date when data were downloaded
- Genomic relationship matrix construction (if applicable)
-
One-step or two-step model
- If the latter, preliminary individual trial analysis methods
- Prediction model training
Developing prediction algorithms
- There will be no attempt to determine if AI has been used for coding. In fact, we will probably (no promise) set up a chatBot to answer questions about functions for programmatic access to T3.
- Prediction algorithms need not limit themselves to data on T3/Wheat, though only publicly available data should be used. Weather data available online could, for example, be useful.
- These trials come from public-sector wheat breeding programs in the United States. If you (as a contestant) happen to work at a program that contributed a test trial, you probably have access to the phenotypes for that trial. On the honor system, you should not use your access to that data while developing your prediction algorithm.
- We will use the CV0_Trials.csv, CV00_Trials.csv, CV0_Accessions.csv, and CV00_Accessions.csv files to verify that appropriate training data is being used. On the honor system, we will assume you are correctly populating those files.
- We welcome submissions from Teams. For such submissions, the roles of each team member should be briefly described in the methods document.
Follow up
Following the competition, a manuscript describing the competition process and the algorithms used will be submitted to G3. All contestants are welcome to be co-authors on that manuscript. Writing assignments and author order will be decided following the March 13th, 2026 deadline.
Questions?
Use the T3/Wheat Contact Us Form. Include the word "Predictathon" in the Subject line.